WP3: Theory and modelling
WP3 Theory and modelling (Researcher co-I) builds new complexity onto state-of-the-art biophysical LLPS theory to construct a connected coarse-grained suite of in silico and theoretical modelling tools that guide, and are informed by WP1 and WP2, building to a final modelling framework for understanding and control of pyrenoid self-aVVePbO\. WP3¶V cRUe RXWSXW LV a PRdeO toolkit for natural and synthetic pyrenoids, parameterised for different species and protein, lipid and starch components. Reciprocally, phenomena and quantitative predictions from WP3 will suggest and inform the detail of experiments in WP1 & 2.
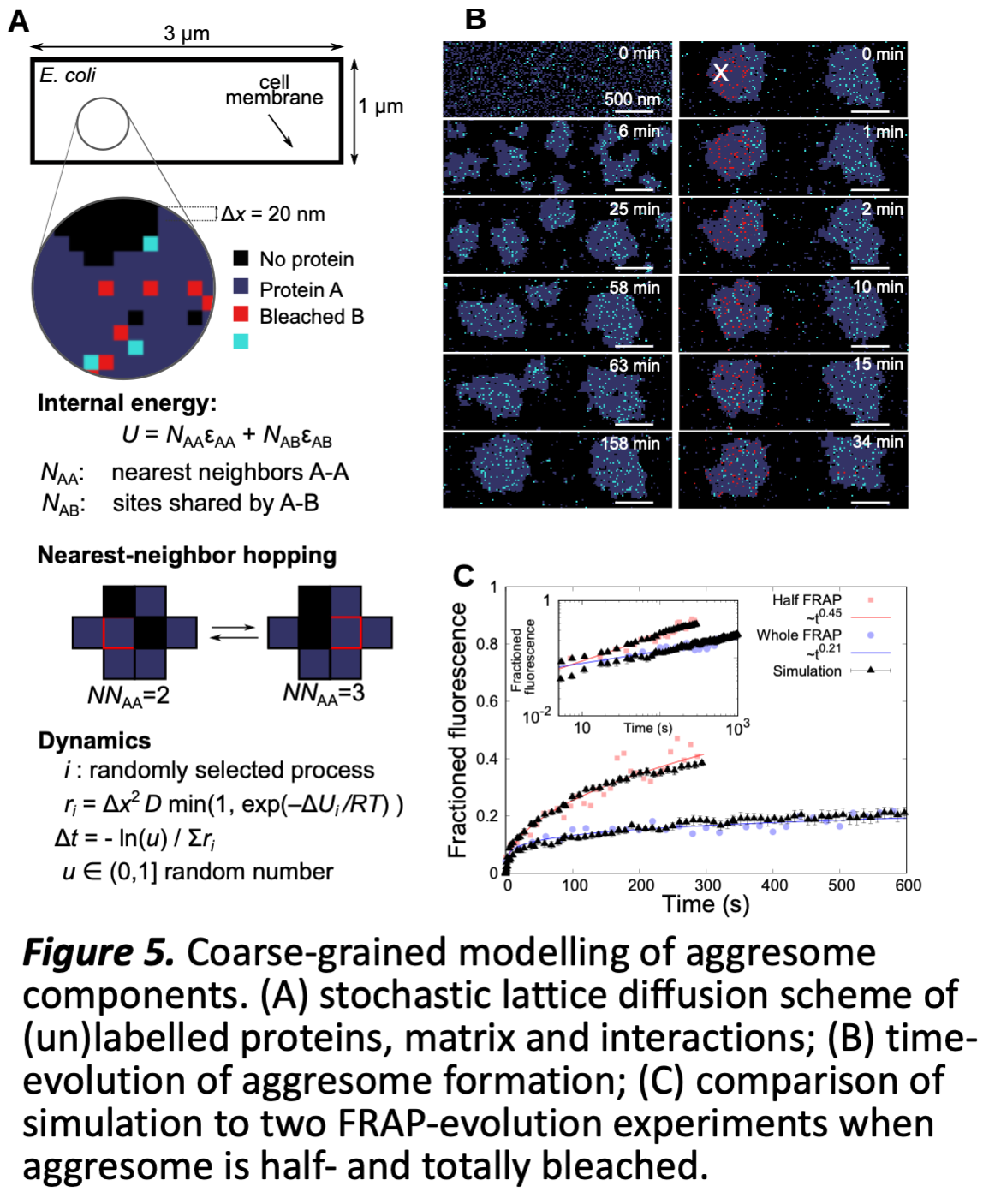
WP3 Preliminary work LQ TM¶V group (R-CoI) has developed a Brownian coarse-grained lattice simulation tool that has successfully modelled simultaneously the dynamics of single proteins, FRAP measurements of diffusion, and LLPS growth, in condensate aggresomes (Fig 5 [10]), demonstrating the ability to model protein association and interpret quantitative and qualitative data and engage with biology and biophysics teams of WP1/2, including skills-transfer via the modelling tools. The modelling inspired individual diffusion measurements of each key protein, exploring dynamic heterogeneity, while allowing interpretation of qualitative in vivo data in terms of quantitative protein SaUaPeWeUV. IW aOVR VXJJeVWed WKe e[LVWeQce Rf µdULYeU¶ aQd µSaVVeQJeU¶ SURWeLQV LQ LLPS fRUPaWLRQ. Simple preparatory lattice-models for tubular lipid thylakoids suggest a role in co-localising proteins at negative curvature. We have also developed theoretical physics tools for non-equilibrium LLPS [13], and finer-scale protein dynamics [14].
The methodology of WP3 adopts both 2D and 3D coarse-grained lattice simulations as its core, but developed in two essential, novel, and distinctive ways: (1) increased functionality from protein only, by degrees, to the higher complexity of structure of lipids and starch through the pyrenoid system hierarchy; (2) selectively-linked to finer levels of modelling supported by MARTINI [15] combined atom models, fully-molecular simulation, combined with models for active, non-equilibrium LLPS.
WP3 Programme of work
WP3.1 Polymeric multivalent linker components (M1- M12): engages WP1.2 and WP2.1 by lattice Brownian simulations of coarse-grained models for Rubisco and EPYC1 (and other linker proteins) parameterised for the systems selected for in vitro phase-space
mapping. Comparison with in vitro two-component data on interaction kinetics and protein diffusion (WP2) will overdetermine the models at each stage.
WP3 will extend the lattice simulation tool tested in preliminary work (above) with resolved protein domain interactions and selectively check protein protein interaction coarse-graining parameterisations through driven molecular simulation (AMBER) and
mid-range coarse-graining (MARTINI).
WP3.2 Internal networks of tubular lipid membranes (M7-M18) off-lattice simulation, including coarse grained lipid thylakoids to inform the adoption of lipids into the fast, large, lattice simulations [7], and incorporation of actively maintained proton-gradients
across the membranes. The models will be parameterised by local curvature-concentration coupling and pH field dynamics, to explore parameter ranges in which tubular/laminar structures form. The results will be compared with in vitro and in vivo data (WP2) on pyrenoid formation and time-ordering of protein and lipid-structure in WT and mutant systems. Selected protein-lipid coarse graining parameterisations will be checked through the finer-scale simulation tools.
WP3.3 Droplet-surface plate structures (M18-M22) will add lattice and off-lattice coarse-grained simulation of starch platelets and associated linker-proteins into (a) protein system alone, (b) protein plus lipid system. Both levels of model will be compared with in vitro and in vivo data (WP2.2) on kinetics and structure of starch sheath formation on artificial and biological pyrenoids
WP3.4 Triggered kinetics of division (M23-M28) will employ the simulation toolkit to identify trigger sites (effective interaction energies) for pyrenoid formation and dissolution, in tandem with in vitro (WP2.3) and in vivo (WP1.4) experiments on triggers, kinetics, (sub-)structure evolution and component dynamics.
WP3.5 Develop simulation toolkit (M29-M36) will combine the previous modelling components at lattice-level, parameterised as required by selected molecular simulation, and coarse-graining the appropriate pathways of driven 'active' phase-separation. A resulting modelling toolkit will focus on the impact pathway of providing an instruction manual for building a synthetic pyrenoid.